Human three-dimensional engineered muscle tissue characterization and intramyocellular lipid modeling
1 Cardiovascular-Renal Metabolic Diseases Research Department,
Boehringer Ingelheim Pharmaceuticals, Inc, Ridgefield, CT, USA
2 Department of Biochemistry and Molecular Biology, University of
Miami, Miller School of Medicine, FL, USA
3 Computational Biology and Digital Sciences Department, Boehringer
Ingelheim Pharmaceuticals, Inc, Ridgefield, CT, USA
4 Material & Analytical Sciences Department, Boehringer Ingelheim
Pharmaceuticals, Inc, Ridgefield, CT, USA
5 Nonclinical Safety & DMPK Department, Boehringer Ingelheim
Pharmaceuticals, Inc, Ridgefield, CT, USA
Originally Published in: Journal of Tissue Engineering (September 2025)
Summary
Enabling Unique Insights into 3D Skeletal Muscle Modeling with Mantarray™
The recent study, "Human three-dimensional engineered muscle tissue characterization and intramyocellular lipid modeling," by Cao et al. (Boehringer Ingelheim) leverages Curi Bio’s Mantarray platform along with Curi Bio’s cells and media to achieve unprecedented insights into human skeletal muscle physiology and metabolic disease in vitro.
The Mantarray system proved to be an invaluable tool in this study, providing:
Quantitative Functional Assessments: Using Mantarray, these researchers longitudinally tracked twitch and tetanic force, key performance measurements of muscle function, in engineered muscle tissues (EMTs), providing physiological insights which are unattainable with 2D culture systems.
Pharmacological Responsiveness: Treating the EMTs with common drugs, like caffeine (which boosted force) and verapamil (which caused force to drop), proved these tissues react functionally like real muscle, validating the Mantarray platform as a reliable approach to screen for new medicines.
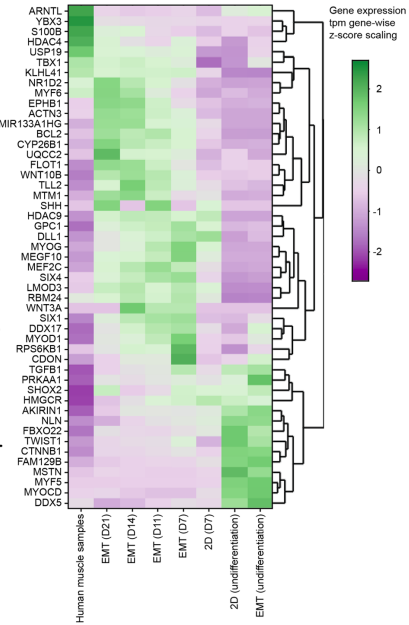
This study further underscored the importance of integrating bulk RNA-sequencing with functional readouts. By using transcriptomic profiling and analysis of fiber type-specific genes, they showed:
Maturation Validation: Gene profiling revealed that hP-Myo EMTs more faithfully recapitulated transcriptional profiles of adult skeletal muscle when compared to hiPS-Myo EMTs which primarily expressed embryonic muscle markers.
Metabolic Modeling: RNA-sequencing was key to validating their intramyocellular lipid (IMCL) model. When the muscle tissues were overloaded with fat to mimic IMCL, the gene data showed a clear disruption in how the cells handled lipids and a shift toward upregulated respiratory electron transport pathways following lipid overload.
These findings not only validate hP-Myo EMTs as a robust platform for physiological studies but also highlight hiPS-Myo EMTs as a tractable system for modeling metabolic abnormalities like IMCL accumulation. A huge step forward for drug screening and personalized therapeutic development of metabolic diseases.